My Work at the Institute for Dynamics of Complex Systems
PhD
Currently, I am completing my doctoral research under the guidance of Prof. Dr. Stefan Klumpp. My goal is to construct a minimal model of the large variety of cellular components, and simulate a significant portion of a whole cell in order to study the extent of liquid liquid phase separation.
Structures formed by 3 patch, patchy particles. This is analogous to a biomolecular condensate. Red particles are the “cores”, yellow particles are “patches”
Structures formed by 2 patch, patchy particles. This is analogous to a linear filaments. Here the color is from a clustering algorithm to distinguish between strands.
Life forms very complex structures, even on the smallest scales. We know individual molecules are not alive but somehow a collection of molecules is. One crucial aspect of life is its ability to self organize, and in a universe where entropy is always increasing, this is fundamental to being alive. Molecules in the cell can form a wide variety of structures, rigid like the cytoskeleton, or fluid like the endoplasmic reticulum. Biomolecular condensates are liquid nano-droplets known to form via liquid liquid phase separation, particularly with bio-polymers.
I am using a minimal model, (in the sense of using the least distinguishing features of the molecules) called the patchy particle model, to study these structures in the cell. The model describes every particle as a core, and any important interactions are mediated through small patches placed on the surface of the cores. Stay tuned for more updates!
Master’s
I wrote my master’s thesis under the supervision of Stefan Klumpp at the Institute for Dynamics of Complex Systems. After progressing to the PhD portion of the Matter to Life program, I chose to continue working in the Klumpp Group. In this blog post, I hope to share the work I got to do with Stefan, and the lovely group of scientists he gathered.
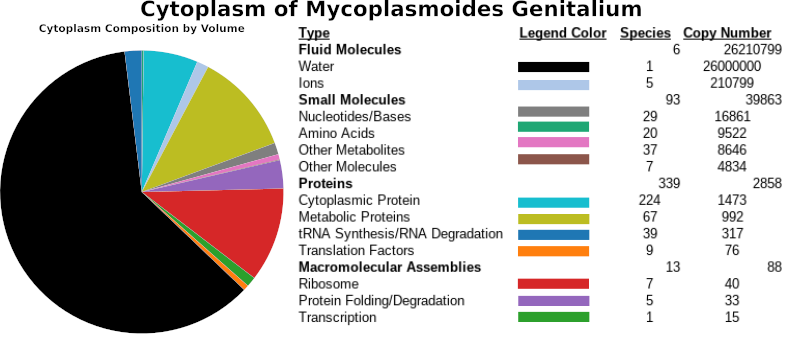
JCVI-syn3A Minimal Cell
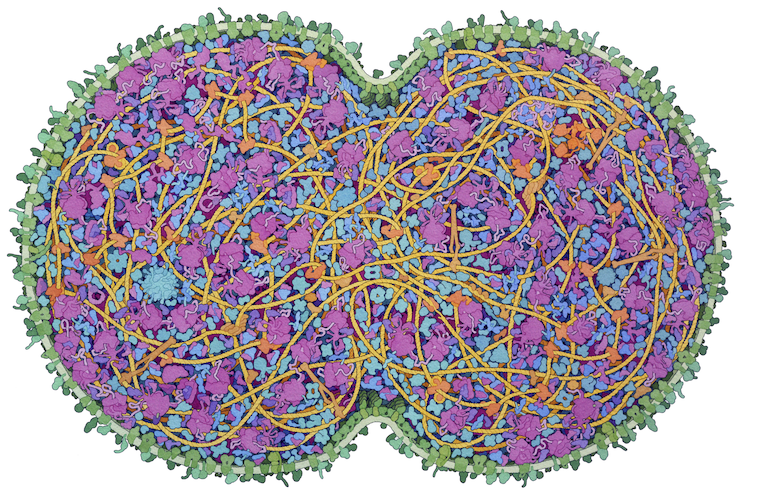
A picture of the components of a bacteria. From David S. Goodsell, RCSB Protein Data Bank.
Right after the masters’ Lab Rotations (Which I talk more about here), I started my work with Stefan Klumpp. Inspired by the measurements I took, and the discourse in the liturature, I chose to continue studying diffusion in the cytoplasm. Others had reported both super- and sub- diffusion of molecules in a variety of settings, which to me was very unexpected, because Brownian motion leading to diffusion as described by the Stokes-Einstein relation, seemed so fundamental. In Stefan’s Group, I was able to return to using simulations to study the problem.
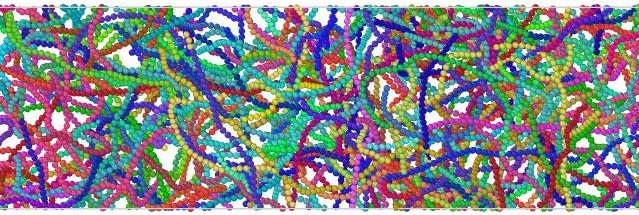
From My Masters Thesis. The components of the cell I included in my biggest simulations
During my Master’s I was granted a lot of freedom in designing my project, and I am very thankful for that learning opportunity. I chose to model a very complex system, the whole cell, and thus I had to model it with a very simple model. As a physicist I imagined the information contained in the sizes, charges, and specific interactions of components in the cell would be enough to see some of the complicated structures seen in life. I combined well studied potentials to model these effects, and created trajectories using a Brownian dynamics integrator. It felt good to be doing molecular dynamics again (context here). Back to the main question of diffusion in the cytoplasm, my model showed a wide variety and distribution of nano-scale clusters, which suggested to me that many cellular components may not be freely diffusing.